defringe¶
Guides & Examples:
Table of File Inputs/Outputs for the Defringe Tools¶
Tool |
Input(s) |
Output(s) |
|---|---|---|
normspflat |
If do_cal is True:
[flat_file]_raw.fits,
[sci_file]_wav.fits
If do_cal is False:
[flat_file]_crj.fits (G750L)
or [flat_file]_sx2.fits (G750M)
[sci_file]_wav.fits
|
[flat_file]_nsp.fits
|
prepspec |
[sci_file]_raw.fits
|
Standard Calstis Outputs:
[sci_file]_flt.fits
[sci_file]_crj.fits
[sci_file]_sx1.fits
[sci_file]_sx2.fits
|
mkfringeflat |
[flat_file]_nsp.fits
[sci_file]_crj.fits (G750L) or
[sci_file]_sx2.fits (G750M)
|
[flat_file]_frr.fits
|
defringe |
[flat_file]_frr.fits
[sci_file]_crj.fits (for G750L)
or [sci_file]_sx2.fits (for G750M)
|
[sci_file]_drj.fits (G750L)
or
[sci_file]_s2d.fits (G750M)
|
Effectiveness of the Defringing Tools¶
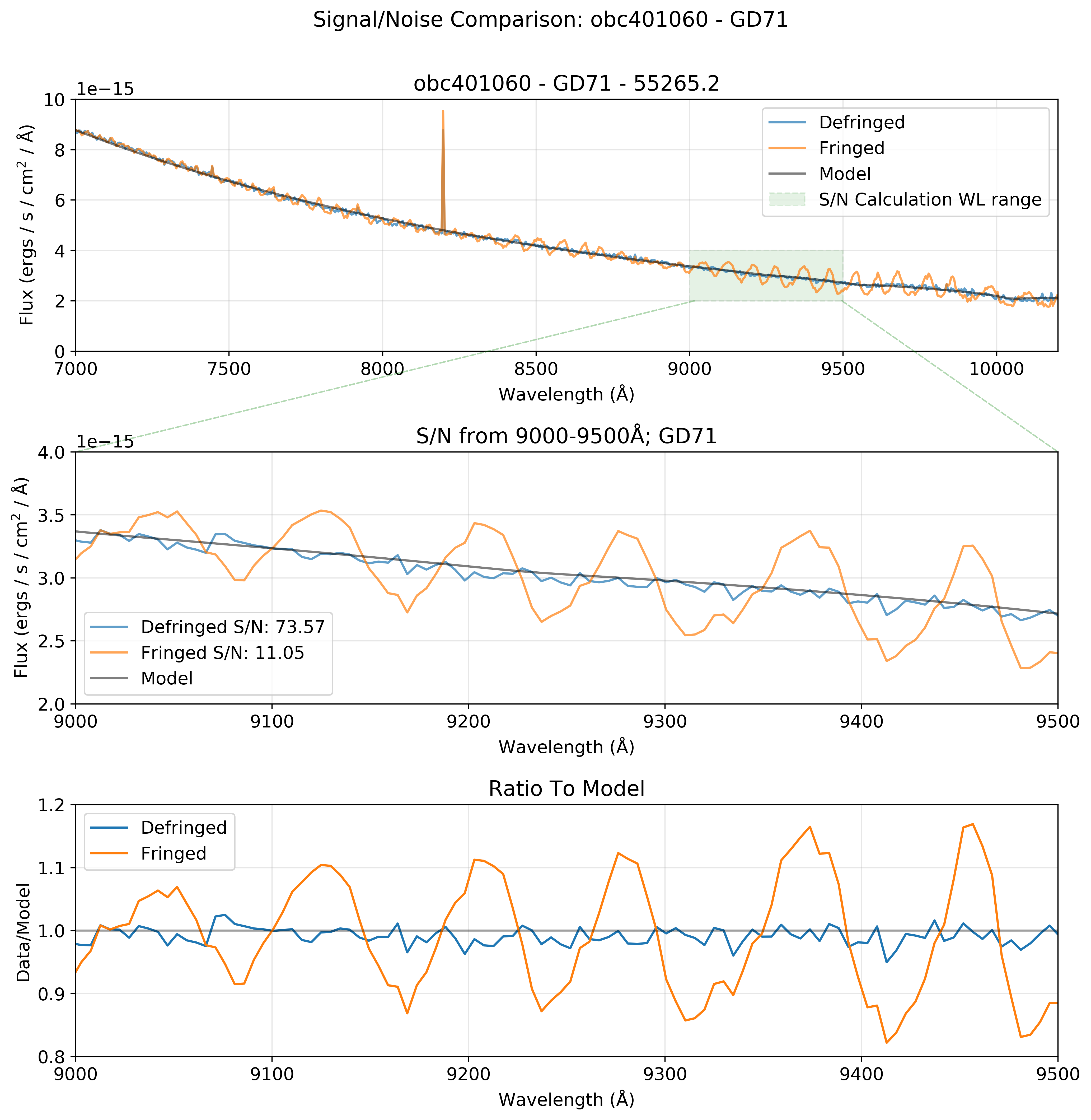
The defringing process has the potential to significantly increase the signal-to-noise ratio of an observation. By removing fringes in an observation of a standard white dwarf target with a well characterized model, we measured an increase in the signal-to-noise ratio from 11 to 73 in the 9000-9500Å wavelength range.
Depending on wavelength range, target, and other observational parameters, we expect that the presence of fringes in an observation may reduce signal-to-noise by up to a factor of 7x. By removing these fringes, we improve the signal-to-noise ratio accordingly.
We calculate the signal-to-noise as 1 divided by the standard deviation of the ratio of the data to the model for a given wavelength range.
Routines¶
prepspec— Calibrate STIS CCD G750L or G750M spectrum before defringingnormspflat— Normalize STIS CCD fringe flatmkfringeflat— Match fringes in STIS fringe flat to those in science datadefringe— Defringe by dividing the science spectrum by the fringe flat
Note
See Section 3.5.5 of the STIS Data Handbook (DHB) for more details on the defringing process.
Warning
These routines are based on PyRAF stsdas.hst_calib.stis defringing tasks, though users should expect numerical discrepancies between these two implementations.
prepspec¶
- stistools.defringe.prepspec(inspec, outroot='./', darkfile=None, pixelflat=None, initguess=None)¶
Calibrate STIS CCD G750L or G750M spectrum before defringing.
Based on the PyRAF stsdas.hst_calib.stis.prepspec task.
- Parameters:
- inspec: str
Name of input 'raw' science spectrum
- outroot: str
Root for output file name. (Default='./')
- darkfile: str or None
Name of superdark image. If None, use DARKFILE in main header of input spectrum.
- pixelflat: str or None
Name of pixel-to-pixel flat. If None, use PIXELFLAT in main header of input spectrum.
- initguess: str or None
Method for initial value estimate for
ocrreject: {None, 'minimum', 'median'}. (Default=None; Use the value in the CRREJTAB.)
- Returns:
- outname: str
Fully qualified name of prepared spectrum (CRJ or SX2 file)
normspflat¶
- stistools.defringe.normspflat(inflat, outflat='.', do_cal=True, biasfile=None, darkfile=None, pixelflat=None, wavecal=None)¶
Normalize STIS CCD fringe flat.
Based on the PyRAF stsdas.hst_calib.stis.normspflat task.
- Parameters:
- inflat: str
Name of input fringe flat
- outflat: str
Name of normalized fringe flat output or directory location. (Default=".")
- do_cal: bool
Perform bias and dark subtraction and CR rejection? (Default=True)
- biasfile: str or None
Name of superbias image. If None, use BIASFILE in main header of the inflat.
- darkfile: str or None
Name of superdark image. If None, use DARKFILE in main header of the inflat.
- pixelflat: str or None
Name of pixel-to-pixel flat. If None, use PFLTFILE in main header of the inflat.
- wavecal: str or None
Name of wavecal file [ONLY FOR G750M SPECTRA]. If None, use WAVECAL in main header of the inflat.
- Returns:
- outname: str
Fully qualified name of the outflat
mkfringeflat¶
- stistools.defringe.mkfringeflat(inspec, inflat, outflat, do_shift=True, beg_shift=-0.5, end_shift=0.5, shift_step=0.1, do_scale=True, beg_scale=0.8, end_scale=1.2, scale_step=0.04, extrloc=None, extrsize=None, opti_spreg=None, rms_region=None)¶
Takes an input science spectrum and a fringe flat that has been normalized using the task
normspflat. The fringe flat is shifted and scaled to produce the minimum RMS when divided into the science data.Based on the PyRAF stsdas.hst_calib.stis.mkfringeflat task.
In
mkfringeflat, the user can specify a range of shifts and scales for the routine to test creating an optimal fringe flat.mkfringeflatwill go through the shift and scale dimensions separately and calculate the RMS using the following steps:For each shift step, apply the shift to the input flat field
Divide the science data by the shifted flat
Divide out the large-scale SED from the science image using a spline fit in order to isolate the fringing pattern (this is called the response image)
Sum the response image along the columns within the RMS region
Calculate the mean and standard deviation of the summed columns of the response image
The RMS value for that shift is given by the standard deviation divided by the mean found in step 5
Fit the RMS values with a quadratic polynomial weighted by the inverse RMS to find the optimal RMS value
Apply the best shift determined in step 7 to the data and repeat steps 1-7 with the scale values to find the best scaling
The RMS values are printed out for each scale and shift but the final best shift and best scale values do not necessarily correspond to the printed values. This is because the routine is calculating the RMS values based on a fit of the data at each scale and shift, rather than being calculated at each discrete step.
- Parameters:
- inspec: str
Name of input science spectrum datafile
- inflat: str
Name of input fringe flat file (usually the output from
normspflat)- outflat: str
Name of output fringe flat to be used in the defringe task
- do_shift: bool
Controls whether the shift between fringe flat and science data is to be calculated
- beg_shift: float
Initial shift to apply to fringe flat
- end_shift: float
Final shift to apply to fringe flat
- shift_step: float
Step-size between shifts to be applied to fringe flat
- do_scale: bool
Controls whether the scaling between fringe flat and science data is to be calculated
- beg_scale: float
Initial scaling to apply to fringe flat
- end_scale: float
Final scaling to appply to fringe flat
- scale_step: float
Step-size between scaling values to be applied to fringe flat
- extrloc: float or None
Extraction location. If set to None, this will be calculated by parabolic interpolation of the peak of the cross-dispersion spectral sum
- extrsize: float or None
Extraction size in pixels. If set to None, this will be set to a reasonable value by this routine
- opti_spreg: list or array-like or None
A list or array representing the section to be used in normalizing the spectrum of the science target before it is divided by the shifted/scaled fringe flat. If set to None, a reasonable range is chosen by this routine. Should be specified like a Python slice, zero indexed.
- rms_region: list or array-like or None
A list or array representing the section to be used in the rms calculation. If set to None, a reasonable range is chosen by this routine. Should be specified like a Python slice, zero indexed.
defringe¶
- stistools.defringe.defringe(science_file, fringe_flat, overwrite=True, verbose=True)¶
Defringe by dividing the science spectrum by the fringe flat.
Based on the PyRAF stsdas.hst_calib.stis.defringe task.
- Parameters:
- science_file: str
The name of the input science file.
- fringe_flat: str
The name of the input fringe flat file. This is the output from
mkfringeflat.- overwrite: bool
The name of the output file will be constructed from the name of the input science file (science_file) by replacing the suffix with 'drj' or 's2d'. If the input name are the same a RuntimeError will be raised, rather than modifying the input in-place. If there is an existing file with the same name as the output name, the existing file will be overwritten if overwrite is True (the default is True).
- verbose: bool
If True (the default), print more info.
- Returns:
- drj_filename: str
The name of the output file. This will have suffix '_drj' if the input is G750L data, and the output name will have suffix '_s2d' if the input is G750M.